Hierarchical Clustering Example¶
Visualize hierarchical clusters
import os.path
import pandas as pd
from time import time
import requests
from graphviz import Digraph
dataset_name = "20_newsgroups_3categories" # see list of available datasets
BASE_URL = "http://localhost:5001/api/v0" # FreeDiscovery server URL
0. Load the example dataset¶
url = BASE_URL + '/example-dataset/{}'.format(dataset_name)
print(" GET", url)
input_ds = requests.get(url).json()
# To use a custom dataset, simply specify the following variables
data_dir = input_ds['metadata']['data_dir']
dataset_definition = [{'document_id': row['document_id'],
'file_path': os.path.join(data_dir, row['file_path'])}
for row in input_ds['dataset']]
Out:
GET http://localhost:5001/api/v0/example-dataset/20_newsgroups_3categories
# 1. Feature extraction (non hashed)¶
1.a Load dataset and initalize feature extraction
url = BASE_URL + '/feature-extraction'
print(" POST", url)
fe_opts = {'max_df': 0.6, # filter out (too)/(un)frequent words
'weighting': "ntc",
}
res = requests.post(url, json=fe_opts).json()
dsid = res['id']
print(" => received {}".format(list(res.keys())))
print(" => dsid = {}".format(dsid))
Out:
POST http://localhost:5001/api/v0/feature-extraction
=> received ['id']
=> dsid = 94bcddd3a1404c32
1.b Run feature extraction
url = BASE_URL+'/feature-extraction/{}'.format(dsid)
print(" POST", url)
res = requests.post(url, json={'dataset_definition': dataset_definition})
Out:
POST http://localhost:5001/api/v0/feature-extraction/94bcddd3a1404c32
2. Calculate LSI¶
url = BASE_URL + '/lsi/'
print("POST", url)
n_components = 100
res = requests.post(url,
json={'n_components': n_components,
'parent_id': dsid
}).json()
lsi_id = res['id']
print(' => LSI model id = {}'.format(lsi_id))
print((' => SVD decomposition with {} dimensions '
'explaining {:.2f} % variabilty of the data')
.format(n_components, res['explained_variance']*100))
Out:
POST http://localhost:5001/api/v0/lsi/
=> LSI model id = bde56bf2ed0943c4
=> SVD decomposition with 100 dimensions explaining 22.26 % variabilty of the data
3. Document Clustering (LSI + Birch Clustering)¶
3.a. Document clustering (LSI + Birch clustering)
url = BASE_URL + '/clustering/birch/'
print(" POST", url)
t0 = time()
res = requests.post(url,
json={'parent_id': lsi_id,
'n_clusters': -1,
'min_similarity': 0.55,
#'max_tree_depth': 3,
}).json()
mid = res['id']
print(" => model id = {}".format(mid))
print("\n4.b. Computing cluster labels")
url = BASE_URL + '/clustering/birch/{}'.format(mid)
print(" GET", url)
res = requests.get(url,
json={'n_top_words': 3
}).json()
t1 = time()
print(' .. computed in {:.1f}s'.format(t1 - t0))
data = res['data']
print(pd.DataFrame(data))
Out:
POST http://localhost:5001/api/v0/clustering/birch/
=> model id = 5c4210bdf1dc43c8
4.b. Computing cluster labels
GET http://localhost:5001/api/v0/clustering/birch/5c4210bdf1dc43c8
.. computed in 0.9s
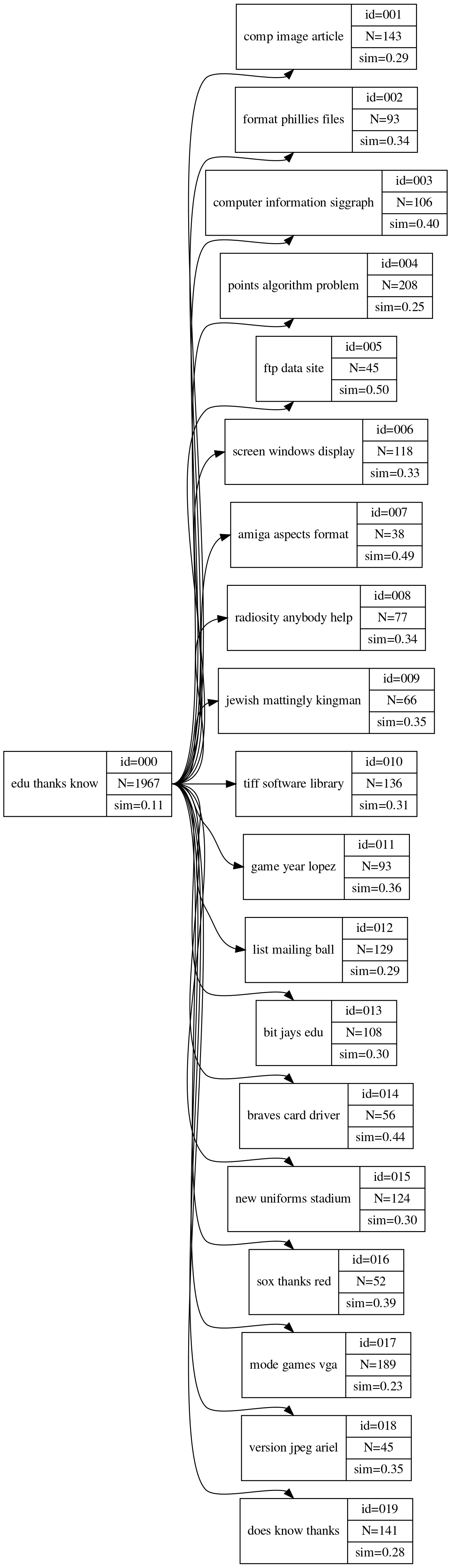
children cluster_depth cluster_id cluster_label cluster_similarity cluster_size documents
0 [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14... 0 0 edu thanks know 0.108 1967 [{'document_id': 225, 'similarity': 0.11136850...
1 [] 1 1 comp image article 0.288 143 [{'document_id': 225, 'similarity': 0.21640175...
2 [] 1 2 format phillies files 0.337 93 [{'document_id': 121, 'similarity': 0.37184011...
3 [] 1 3 computer information siggraph 0.397 106 [{'document_id': 100, 'similarity': 0.00270706...
4 [] 1 4 points algorithm problem 0.253 208 [{'document_id': 144, 'similarity': 0.31973796...
5 [] 1 5 ftp data site 0.498 45 [{'document_id': 196, 'similarity': 0.63168856...
6 [] 1 6 screen windows display 0.325 118 [{'document_id': 5329, 'similarity': 0.1170009...
7 [] 1 7 amiga aspects format 0.492 38 [{'document_id': 441, 'similarity': 0.58264766...
8 [] 1 8 radiosity anybody help 0.337 77 [{'document_id': 361, 'similarity': 0.13379389...
9 [] 1 9 jewish mattingly kingman 0.353 66 [{'document_id': 16129, 'similarity': 0.327495...
10 [] 1 10 tiff software library 0.307 136 [{'document_id': 6889, 'similarity': 0.1279159...
11 [] 1 11 game year lopez 0.361 93 [{'document_id': 169, 'similarity': 0.44509086...
12 [] 1 12 list mailing ball 0.294 129 [{'document_id': 17689, 'similarity': 0.097539...
13 [] 1 13 bit jays edu 0.300 108 [{'document_id': 3969, 'similarity': 0.4519637...
14 [] 1 14 braves card driver 0.440 56 [{'document_id': 40401, 'similarity': 0.441856...
15 [] 1 15 new uniforms stadium 0.300 124 [{'document_id': 961, 'similarity': 0.46294582...
16 [] 1 16 sox thanks red 0.392 52 [{'document_id': 15876, 'similarity': 0.630474...
17 [] 1 17 mode games vga 0.228 189 [{'document_id': 0, 'similarity': 0.2483949485...
18 [] 1 18 version jpeg ariel 0.352 45 [{'document_id': 53824, 'similarity': 0.463246...
19 [] 1 19 does know thanks 0.278 141 [{'document_id': 2401, 'similarity': 0.3866424...
3.b Hierarchical cluster visualization
ch = Digraph('cluster_hierarchy',
node_attr={'shape': 'record'},
format='png')
ch.graph_attr['rankdir'] = 'LR'
ch.graph_attr['dpi'] = "200"
for row in data:
ch.node('cluster_{}'.format(row['cluster_id']),
'{{<f0>{}| {{<f1> id={:03} |<f2> N={} |<f3> sim={:.2f} }}}}'
.format(row['cluster_label'],
row['cluster_id'],
row['cluster_size'],
row['cluster_similarity']))
def create_hc_links(node, ch, data):
for child_id in node['children']:
ch.edge('cluster_{}:f2'.format(node['cluster_id']),
'cluster_{}:f0'.format(child_id))
create_hc_links(data[child_id], ch, data)
create_hc_links(data[0], ch, data)
tmp_dir = os.path.join('..', '..', 'doc', 'engine', 'examples')
if os.path.exists(tmp_dir):
ch.render('cluster_hierarchy', directory=tmp_dir, cleanup=True)
else:
ch.view()
- Delete the extracted features
url = BASE_URL + '/feature-extraction/{}'.format(dsid)
requests.delete(url)
Total running time of the script: ( 0 minutes 4.375 seconds)